Congratulations to the Schnitzler Lab on their recent publication in BMC Genomics, titled “An updated and spatially validated somatic single-cell atlas of Hydractinia symbiolongicarpus.”
Co–first authors Drs. Jingwei Song and Danielle de Jong, together with Dr. Justin Waletich (former Schnitzler Lab graduate student) and Dr. Christy Schnitzler, and in collaboration with Dr. Andy Baxevanis (NIH), contributed to the generation and analysis of the data presented in this study.
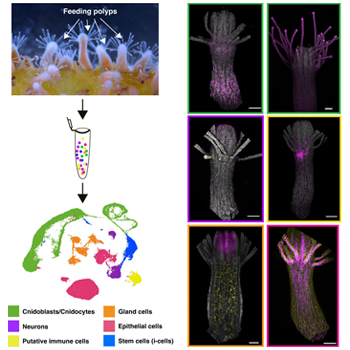
This work presents an updated single-cell atlas of Hydractinia comprising over 47,000 cells. In simple terms, the authors created a detailed map of the individual cells that make up Hydractinia, and the relationship of these cells to each other based on the genes that are expressed. Importantly, the authors didn’t just show this computationally, they also selected marker genes and confirmed the cell types these genes were expressed in, and where these cell types are located within the animal. This data is openly available and provides an invaluable resource for the cnidarian research community. The full paper is available here:
https://link.springer.com/article/10.1186/s12864-025-12201-9
The work is also discussed in an accompanying review on cnidarian sensory systems by Drs. Schnitzler, Song, and de Jong which can be viewed with the following link: https://academic.oup.com/icb/article/65/3/701/8161048
Abstract
Single-cell RNA sequencing (scRNA-seq) has revolutionized transcriptomic research, enabling the creation of detailed tissue, organ, and species-level atlases for model organisms. In Hydractinia, a cnidarian model for stem cell and regeneration studies, recent atlases have revealed key insights into cell types and developmental processes. However, these atlases remain limited in cell numbers and transcriptomic depth and cell type assignments were largely made in silico. Here, we present an updated Hydractinia single-cell atlas by integrating new datasets from fixed cells with previously published live-cell data. This expanded atlas captures over 47,000 cells from feeding polyps and stolon tissue, recovering and refining major somatic cell lineages including cnidocytes, neurons, gland cells, epithelial cells, and stem cells (i-cells), as well as identifying a novel population of putative immune cells. We investigated the spatial expression patterns of selected marker genes and validated all major cell types and several cell states. Our analyses uncovered a previously undescribed neural subtype, two spatially distinct gland cell populations, a stolon-specific cell type, and a putative immune cell cluster. Additionally, we recovered and explored a complete Hydractinia cnidocyte trajectory with two distinct endpoints, supported by spatial marker gene expression that reflects the developmental progression of cnidoblasts as they mature and migrate towards the tentacles. Subclustering of somatic i-cells revealed putative progenitor states and a potential population of true stem cells. Together, this atlas significantly advances our understanding of Hydractinia cellular diversity and dynamics, allowing us to generate new hypotheses and provide a valuable resource for the cnidarian research community and beyond.