VERONICA HINMAN, PH.D.

Veronica Hinman, Ph.D.
Professor of Biology, Director Whitney Laboratory for Marine Bioscience
Dr. Veronica Hinman obtained bachelor's degrees in Mechanical Engineering and Zoology from the University of Queensland in Australia. She completed her PhD at the University of Queensland, where she spent time doing research at the University’s Heron Island Marine Research Station on the Great Barrier Reef. She then worked as a postdoctoral researcher at Caltech with Eric Davidson, studying the evolution of gene regulatory networks (GRNs) using Echinoderm embryos. Her research focuses on GRNs and the evolution of development and regeneration, mainly focused on cis-regulatory mechanisms and comparative functional genomics using echinoderms. She has led a productive, funded research lab at Carnegie Mellon University from 2006-2024. She is the co-director of an NIH-funded resource program in bioinformatics to run Echinobase.org. The core mission of this model organism knowledgebase is to provide gold-standard, curated, and publicly accessible integrated genomics data.


Research Program
Our group has several broad interests that fall under the umbrella of “how genomes control cell fate decisions during development and how this evolves”. We think about development as embryonic and non embryonic (i.e. regeneration). We have ongoing projects in EvoDevo, including cooption, body plan evolution, and cis regulatory evolution, and in Regeneration, including, cell pluripotency and neurogenesis.
Our motto is to not be limited by approaches and technologies; therefore we work in molecular biology, genomics, biochemistry, microscopy, high throughput sequencing, embryology and computational biology. We’re very collaborative because we always want to find out how to do new things and we like working with diverse people. We also work at whatever scale of organization will best answer our questions, and thus we take very systems approaches as well as more reductionist approaches.
Our lab uses a range of echinoderm species as model system. These organisms are especially well suited to high throughput analyses as we can readily generate and raise transgenic embryos and larvae. We work with multiple species of echinoderms, including sea urchins, Stronglyocentrotus purpuratus, Lytechinus variegatus, the sea sea star Patiria miniata and the sea cucumber Parastichopus parvimensis.
Current Projects
The Evolution of novelty and cell type evolution
A significant focus of the lab's work is to understand how GRNs evolve to produce new cell types without compromising organismal viability. A GRN (Gene Regulatory Network) is a collection of molecular regulators, including genes, transcription factors, and signaling molecules, that interact to control gene expression in a cell or organism. GRNs dictate when, where, and how genes are turned on or off, shaping developmental processes, cell differentiation, and evolutionary changes.
One of the active areas of research is the evolution of novel cell types and their underlying gene regulatory networks (GRNs). We use echinoderms, such as sea stars and sea urchins, as they have some very clear examples of novel cell types - including the pigment cells, found only in sea urchins and the larval skeleton, found only in sea urchins and brittle stars. The lab constructs comprehensive gene expression atlases for these species by employing single-cell and single-nucleus RNA sequencing, enabling detailed comparisons of cell type diversity and evolutionary trajectories. This work has shown unexpected findings, for example, the similarities between neural cell types and pigment cell types, which provide new insights into how cell types evolve.
Mechanisms of Regeneration
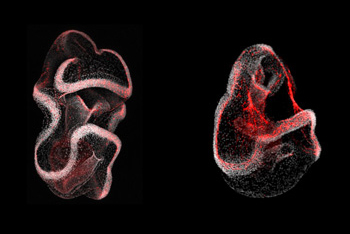
Another lab focus is understanding the mechanisms underlying regeneration. We investigate how echinoderms, particularly sea stars and sea urchins, restore lost body parts, emphasizing the role of gene regulatory networks (GRNs) in this process.
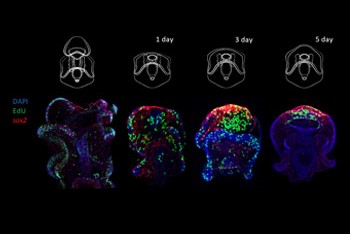
A significant aspect of the work involves studying the regeneration of the nervous system. Our team developed transgenic tools to trace cell lineages during regeneration, revealing that following injury, specific cells can re-specify into neural lineages, contributing to nervous system regeneration. This process involves the activation of specific genes, such as sox2, which plays a crucial role in cell fate determination during regeneration.
Additionally, we have characterized whole-body regeneration in sea star larvae, identifying conserved gene expression changes that occur during the regenerative process. These studies have shown that sea star larvae can proportionally regenerate their bodies after bisection, involving wound-induced gene expression and localized cell proliferation at the injury site.
This research provides valuable insights into echinoderm regeneration's cellular and molecular mechanisms, contributing to a broader understanding of regenerative biology and its evolutionary implications.