 The Whitney Laboratory for Marine Bioscience
The Whitney Laboratory for Marine Bioscience
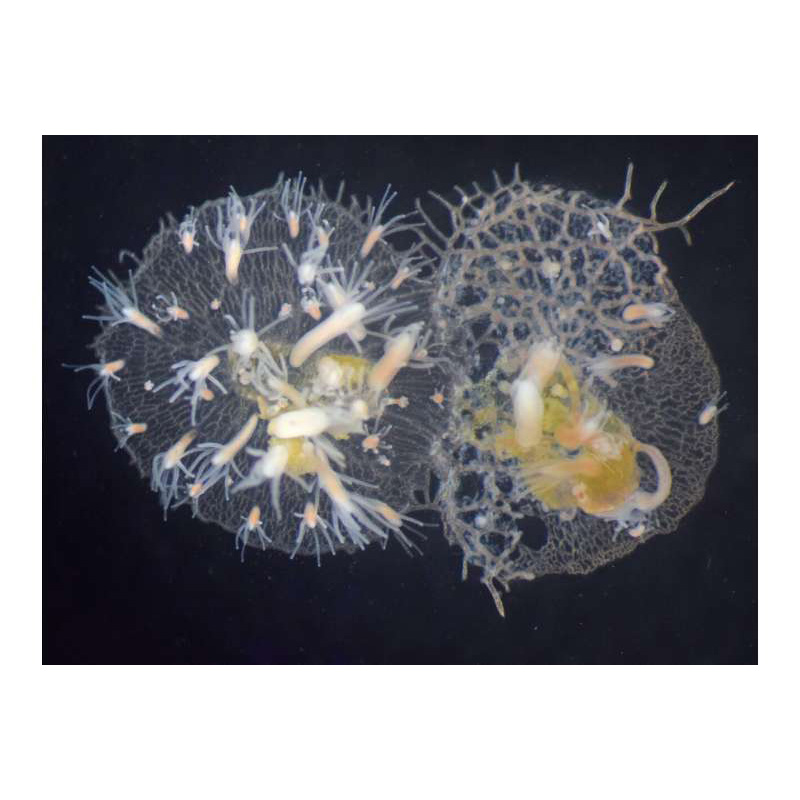
Congratulations to Dr. Christine Schnitzler and her co-authors from the University of Pittsburgh and the National Human Genome Research Institute of the NIH for publishing a new paper in the Proceedings of the National Academy of Sciences (PNAS) – “A family of unusual immunoglobulin superfamily genes in an invertebrate histocompatibility complex”
https://www.pnas.org/doi/10.1073/pnas.2207374119
In this study, researchers focused their attention on a region of Hydractinia’s genome known as the allorecognition region responsible for self/non-self recognition. Hydractinia grow as colonies on the shells of hermit crabs. As the colonies grow and compete for space, they often come in contact with one another. If the colonies recognize each other as “self”, they fuse. If they identify each other as “non-self”, they fight. The researchers involved identified and sequenced a huge family of 41 allorecognition (Alr) genes in the allorecognition region of Hydractinia and showed for the first time that Alr proteins belong to a special group of proteins called the immunoglobulin superfamily (IgSF) – which are important for adaptive immunity in mammals and other vertebrates. This invertebrate allorecognition complex is remarkably similar to the major histocompatibility complex, leukocyte receptor complex, and the NK complex in vertebrates and could shed light on how such complexes evolved and ultimately could help inform our understanding of immune signaling.
Most colonial marine invertebrates are capable of allorecognition, the ability to distinguish between themselves and conspecifics. One long-standing question is whether invertebrate allorecognition genes are homologous to vertebrate histocompatibility genes. In the cnidarian Hydractinia symbiolongicarpus, allorecognition is controlled by at least two genes, Allorecognition 1 (Alr1) and Allorecognition 2 (Alr2), which encode highly polymorphic cell-surface proteins that serve as markers of self. Here, we show that Alr1 and Alr2 are part of a family of 41 Alr genes, all of which reside in a single genomic interval called the Allorecognition Complex (ARC). Using sensitive homology searches and highly accurate structural predictions, we demonstrate that the Alr proteins are members of the immunoglobulin superfamily (IgSF) with V-set and I-set Ig domains unlike any previously identified in animals. Specifically, their primary amino acid sequences lack many of the motifs considered diagnostic for V-set and I-set domains, yet they adopt secondary and tertiary structures nearly identical to canonical Ig domains. Thus, the V-set domain, which played a central role in the evolution of vertebrate adaptive immunity, was present in the last common ancestor of cnidarians and bilaterians. Unexpectedly, several Alr proteins also have immunoreceptor tyrosine-based activation motifs and immunoreceptor tyrosine-based inhibitory motifs in their cytoplasmic tails, suggesting they could participate in pathways homologous to those that regulate immunity in humans and flies. This work expands our definition of the IgSF with the addition of a family of unusual members, several of which play a role in invertebrate histocompatibility.