 The Whitney Laboratory for Marine Bioscience
The Whitney Laboratory for Marine Bioscience

Information for Graduate Students
Dr. Leonid Moroz earned his Ph.D. in physiology and evolutional and developmental biology under the tutelage of Dr. D.A. Sakharov at the Institute of Developmental Biology, Academy of Sciences in Moscow, Russia. His postdoctoral research was done with Dr. William Winlow at the University of Leeds in the UK and with Dr. Rhanor Gillette at the University of Illinois, Urbana. Dr. Moroz was an HHMI International Scholar and, in 1998, was recruited to the University of Florida.
Email: moroz@whitney.ufl.edu
The human brain is the most complex molecular machine known. It is composed of a hundred billion nerve cells, each of which is unique and makes innumerable connections with other nerve cells. Together these cells and all their connections allow us to move, think, dream, imagine and learn.
It would seem that the best and the most efficient way to treat any disease is to understand the normal biology of a physiological process, cell or organ at the level of the genes that make it work. Understanding brain complexity starts with the most fundamental questions: What makes a neuron? What is the molecular “toolkit” needed to make a neuron in the first place? Is there only one or are there many “toolkits” to support the origin and maintenance of neural organization?
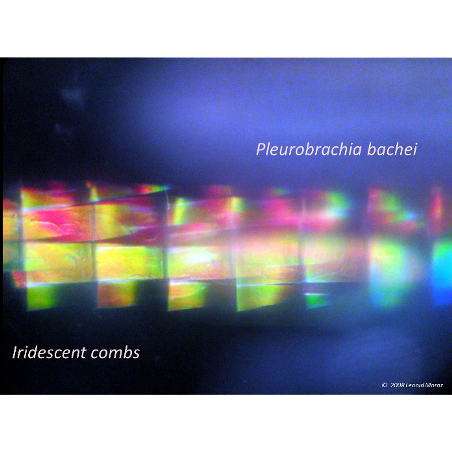
Comparative neurobiology suggests that there are many ways to make a neuron. Astonishingly, our analysis reveals that simplex brains might have independently evolved at least five to seven times during the 550 million years of animal evolution. The hypothesis we are testing is that the complex brains we find in representatives of existing animal phyla are the result of parallel evolution of different ancestral cell lineages. We want to reconstruct how the descendents of these cell lineages “come together” to form a brain in the octopus, or honey bee, or human. We are working on a broad spectrum of animals representing different levels of neural organization and tissue complexity: from the simplest sponges to molluscs (slugs, nautilus and octopus) and arthropods.
Image: Pleurobrachia bachei

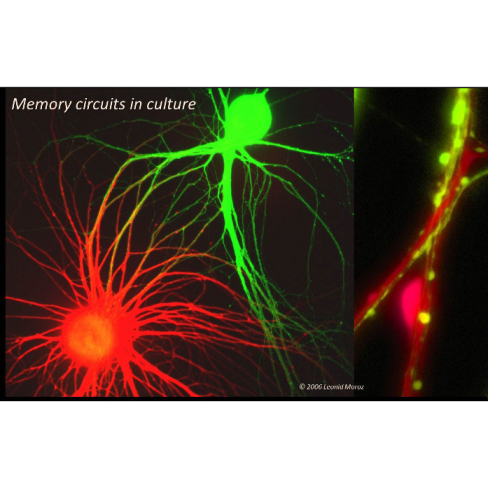
Another research goal is to understand the molecular mechanism of the formation and long-term maintenance of memories. Here, the sea slug Aplysia and related marine molluscs lead the way. Their giant nerve cells allow us to probe the critical molecular events of learning and memory in real time, all the way from genes to behaviors. In fact, we can follow the activity of all the genes in a single neuron as it learns and remembers!
In doing so we are asking questions which are difficult or impossible to address elsewhere. How does the activity of more than 20,000 genes within a single cell grow synapses and form memories that last a lifetime? Is there only one way, or are there multiple ways to do this and why? Since the genes and the way they are regulated in our brain cells are so similar to those in Aplysia, this understanding has tremendous potential application for diseases of the human brain and nervous system.
Our laboratory works to characterize basic mechanisms underlying the design of nervous systems and evolution of neuronal signaling mechanisms. The major questions are: (1) why are individual neurons so different from each other, (2) how do they maintain such precise connections between each other, (3) how does this fixed wiring result in such enormous neuronal plasticity and (4) how does this contribute to learning and memory mechanisms? By taking advantage of relatively simpler nervous systems of invertebrate animal models, we combine neuroscience,genomics, bioinformatics, evolutionary theory, zoology, molecular biology, microanalytical chemistry and nanoscience to understand how neurons operate, learn and remember.
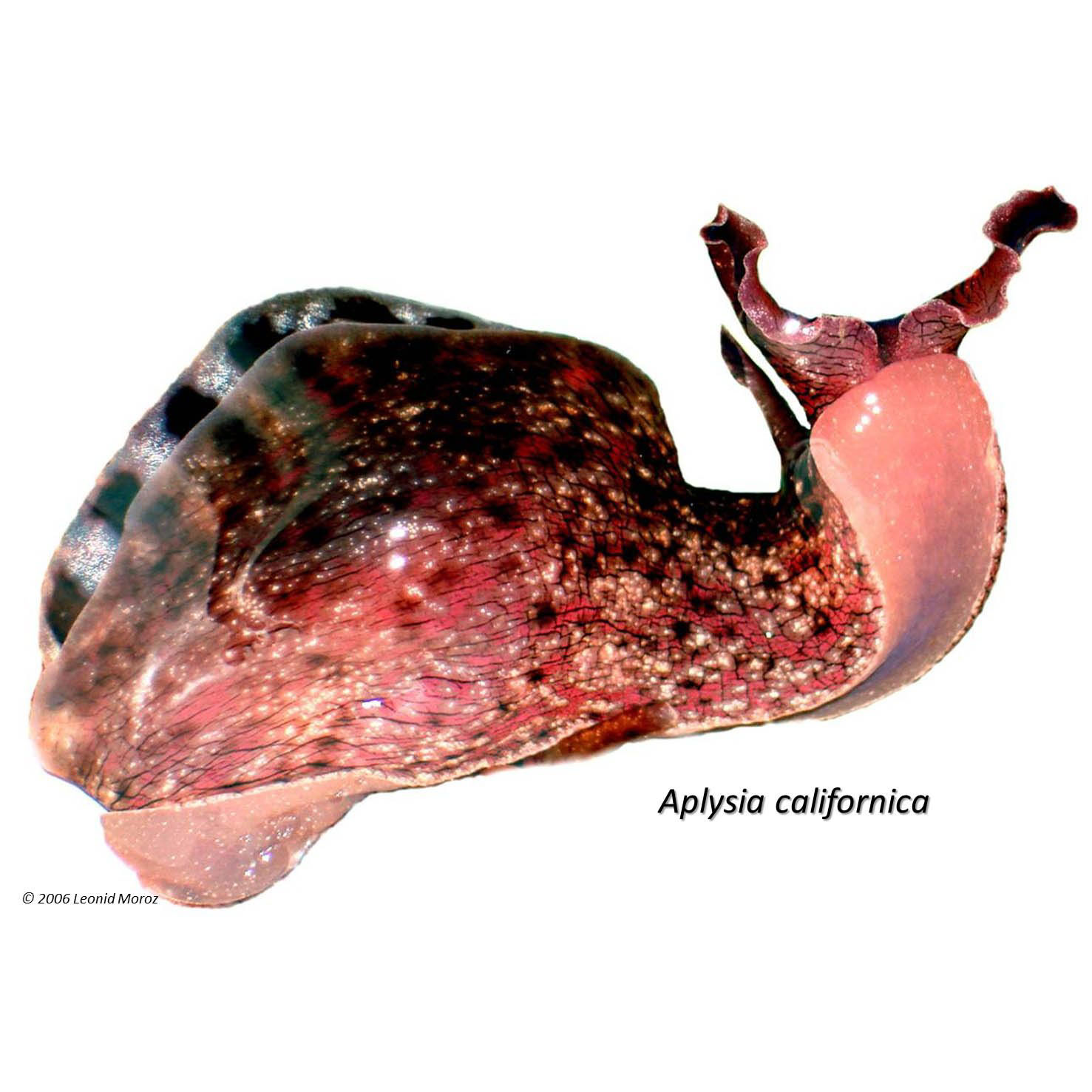
As part of the NIH Center of Excellence in Genomic Sciences, our first project investigates the genomic bases of neuronal identity and plasticity. Due to the tremendous difficulties in mapping single cells and processes in the mammalian brain, we study the giant neurons of the sea slug Aplysia californica, a well-established model organism for cellular neuroscience. Our objective is to investigate nearly all messenger RNA (mRNA) involved in simple feeding and defensive networks.
Genomic Bases of Neuronal Identity and Plasticity. This year we have completed our collection and initial inventory of Aplysia neuronal mRNAs from identified neurons and even from their individual processes and growth cones. In collaboration with Columbia University (J. Ju and E. Kandel), we have generated more than 230,000 molecular markers of expressed genes (ESTs) and identified thousands of unique Aplysia genes operating in the central nervous system. These include genes associated with neurodevelopment, synaptogenesis and memory. We also identified several dozen genes homologous to those associated with human diseases such as Alzheimer’s disease and causes of mental retardation (the Alzheimer’s disease related protein and the Fragile X mental retardation protein).
We are analyzing the functional role of these novel and critical genes using Aplysia-specific microarrays, in situ hybridization, capillary electrophoresis, confocal and time-lapse imaging and microelectrode techniques. These technologies are integrated to elucidate the entire molecular machinery of a living neuron. The neuronal transcriptome of Aplysia has been published. For more information and to view the abstract, click here. Our lab has also embarked on a project, supported by NIH, to sequence the entire Aplysia genome. Click here to find out more.
Development of Innovative Bionanotechnologies for Direct Single Cell Genomic and Microchemical Analysis. We are working to reveal the complex, large-scale molecular machinery of individual neurons by monitoring, at real physiological time, the activity of thousands of genes and hundreds of metabolites in the cell. Our goal is to identify a set of genes that determine neuronal identity and control long-term nervous system modifications such as injury, learning and memory. Although we have succeeded with single neuron gene expression profiling using custom made microarrays, real-time measurements and large-scale metabolite proofing is a desired but not yet achieved goal. As the first step in this direction, we are developing a new generation of intracellular sensors called molecular beacons (in collaboration with J. Ju, N. Turro and W. Tan), for direct imaging of native mRNAs in living neurons and synapses in real physiological time. These new probes allow us to monitor mRNA trafficking and relocation in both cell culture and intact ganglia as neurons learn and remember. We also continue to use microanalytical approaches to detect and measure microscopic quantities of important chemicals in neurons. The primary technique, capillary electrophoresis, is so exquisitely sensitive that less than 0.01 of the volume of a cell is all that is required for a detailed chemical analysis of its contents. Developing (in collaboration with Jonathan Sweedler) and extending these microchemical technologies to other cell types and multiple metabolites (the neuronal “metabolome”) as well as their integration to real-time electrophysiological analysis (by on-line sampling of cytoplasm) are our goals for the coming years.
The third project focuses on physiological mechanisms and evolution of gaseous signaling in the nervous system. Nitric oxide (NO) is a signaling molecule involved in a myriad of biological functions. Nerve cells that produce NO, the most mysterious and least analyzed population of cells in the brain, are now our primary research target. Recently, we identified and mapped these neurons, successfully isolating genes involved in NO synthesis in molluscs, lower chordates and cnidarians. We are now examining how NO acts in neuronal networks. Gaseous transmission has been reconstructed in cell culture where we have started to create a molecular portrait of a nitrergic neuron and identify targets of NO action. The emerging picture is that NO can be highly compartmentalized in a neuron, yet it might affect numerous signaling pathways by direct and indirect interaction with other cellular messengers. Finally, we discovered that in certain neurons NO can be generated by a non-enzymatic mechanism via nitrite-ascorbate interactions, and we continue to characterize this non-traditional mechanism of NO synthesis in cells.
The goal of our fourth project is to use comparative strategies to determine what genes define neurons, focusing upon basal animal groups. First, we make and sequence cDNA libraries from Trichoplax, having the simplest organization of any known animal, with only four distinguishable cell types, but with prominent social behavior. Second, this year we started to sequence Xenoturbella, an enigmatic marine organism that is considered as the representative of the most basal deuterostome group with a nervous system in the form of a nerve net. Finally, we use larval nervous systems from molluscs and echinoderms as important outgroups in this genomic screening. Such an approach will provide a foundation for phylogenetic analysis of gene loss and gain in both basal and more derived nervous systems and identify a subset of neuron-specific genes. These projects are performed in collaboration with Profs. P. Anderson and B. Battelle. The goals of the research are twofold: to establish novel genomic models and to understand the molecular basis and evolution of major signaling pathways focusing on the identification of evolution’s most conserved mechanisms of neuronal identity and plasticity as well as trends leading to innovations in neuronal functions.
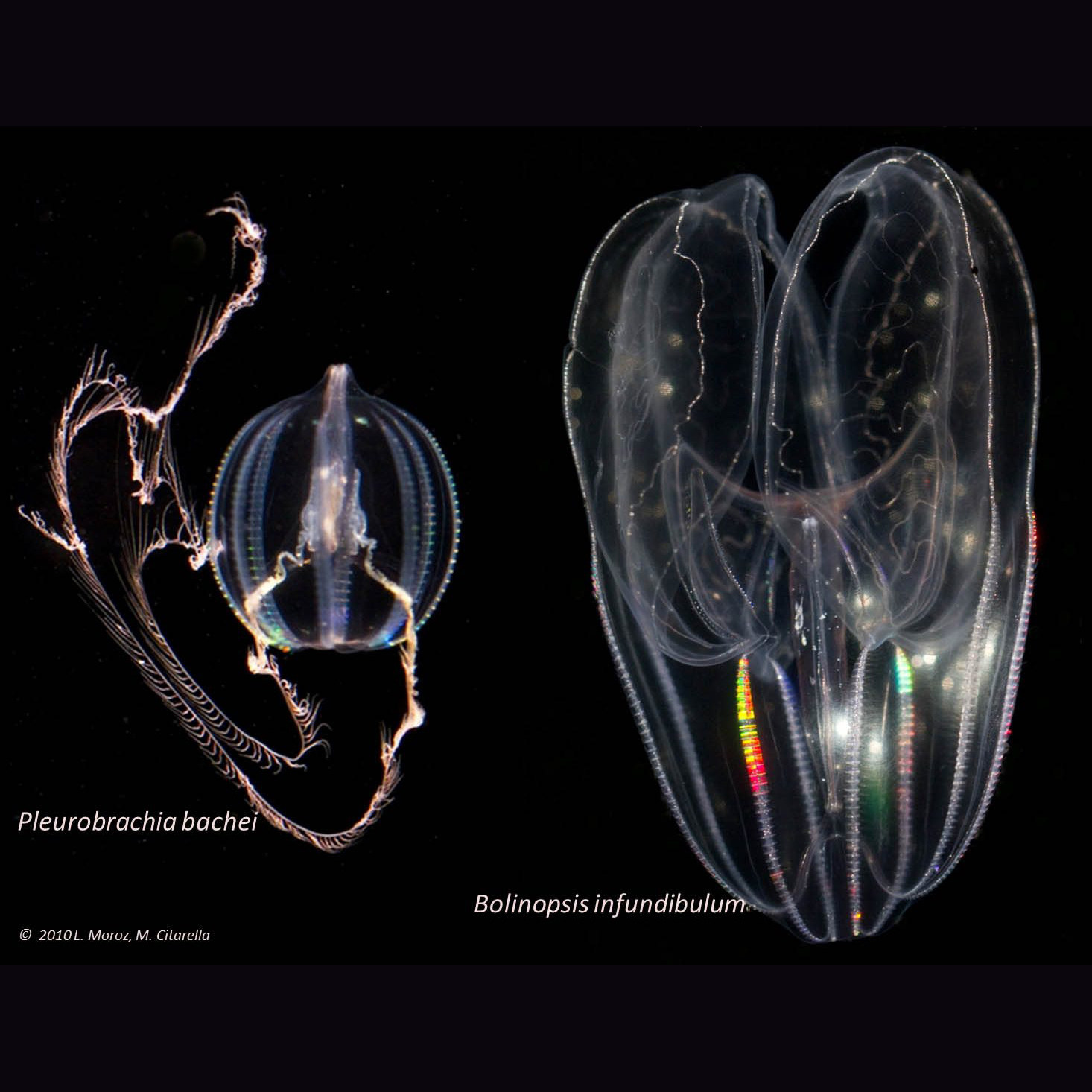
Our laboratory works to characterize basic mechanisms underlying the design of nervous systems and parallel evolution of neural circuits, neuronal signaling mechanisms and brains.
Read More