 The Whitney Laboratory for Marine Bioscience
The Whitney Laboratory for Marine Bioscience
Congratulations to Whitney Laboratory's Dr. Joseph Ryan for his recent publication in Gigascience.
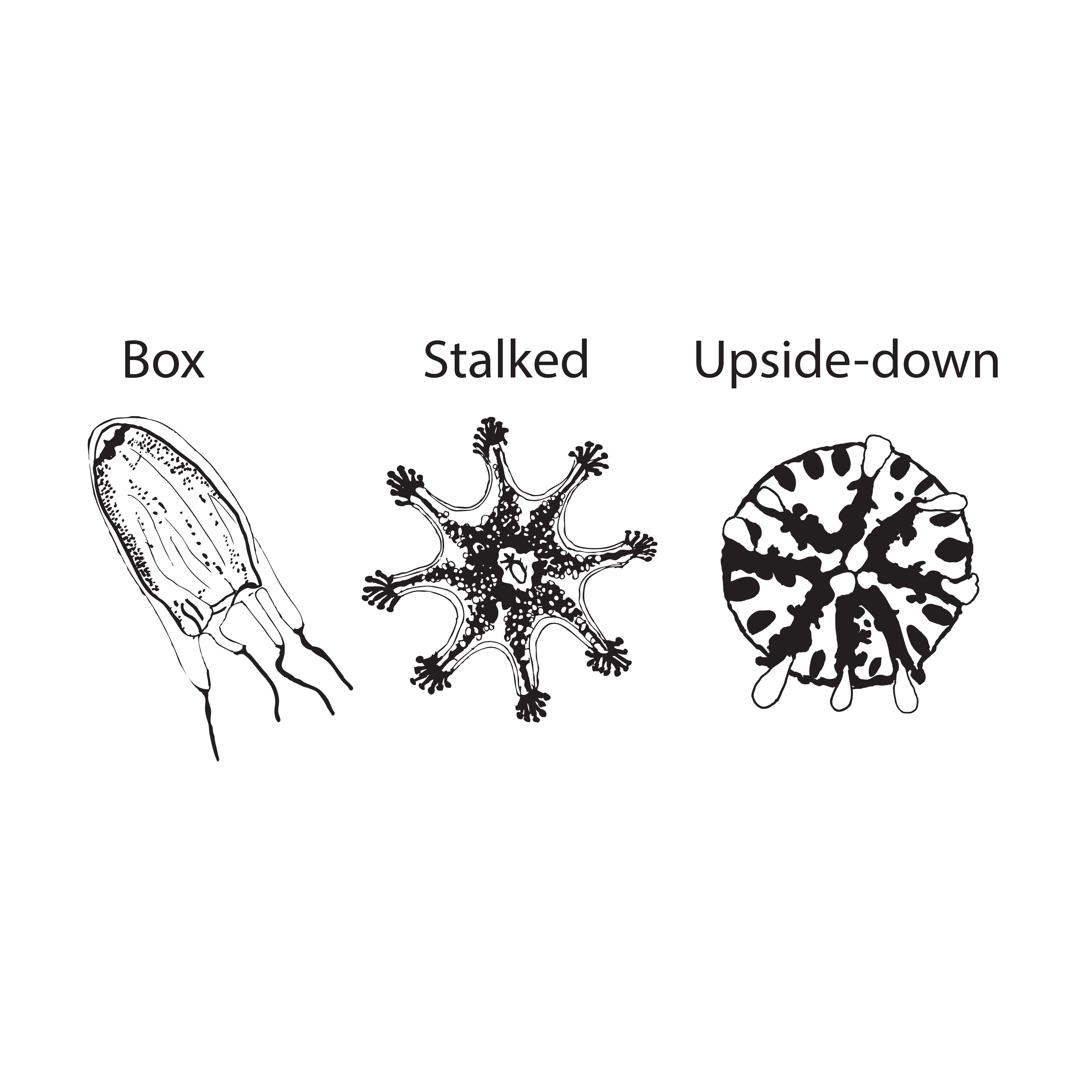
A international collaboration involving 8 institutions led by Joseph Ryan at the Whitney Laboratory describes sequencing and analyses of genomes of three disparate jellyfish species: a box jellyfish (Alatina alata), a stalked jellyfish (Calvadosia cruxmelitensis), and an upside down jellyfish (Cassiopea xamachana). Box jellyfish (Cubozoa) are among the most venomous animals on the planet and therefore their genomes are important resources for developing antivenoms as well as potential drugs. Like vertebrates, box jellies have eyes with image-forming lenses, which makes their genome an important tool for understanding the evolution of vision. Stalked jellies (Staurozoa) attach themselves to hard surfaces with a stalk (like a polyp) instead of having a free-swimming adult phase. The peculiar anatomy of these animals suggests that their genome may offer insight into the evolution of the body plans and life history in jellyfish. The genome of Cassiopea xamachana adds another important comparative data point for understanding these fascinating animals. Since Cassiopea has become a model in a range of scientific fields (e.g., developmental biology, symbiosis research, and behavioral neurobiology) the genome will be an especially important experimental resource.
With these new genomes, for the first time, they were able to compare gene content of all 5 classes of cnidarians (Anthozoa, Hydrozoa, Cubozoa, Staurozoa, Scyphozoa). This broad survey provides an important overview of genomic evolution in cnidarians. Their survey included a focused assessment of venom-related genes, providing insight into the evolution of cnidarian venom. They also showed the utility of these genomes by describing the evolutionary history of a gene-fusion event and genomic linkage of a pair of important developmental genes called homeobox genes. They showed that in both cases these events predate the last common medusozoan ancestor, and therefore, likely represent important genomic constraints that are under purifying selection.
Like those of other animals, jellyfish genomes are highly complex and underlay a huge diversity of biological phenomena, including life cycles, prey preferences, metamorphosis, swimming behaviors, population blooms, sexual behaviors, environmental stress reactions, cell type determination, stinging capsule development and location, etc. These data will be an important resource for future efforts to build molecular markers that can be applied to constructing an accurate and comprehensive tree of life for Cnidaria. They also will be important for accurately inferring gene loss and understanding the evolution of genome architecture (e.g., gene order) in cnidarians. Together, these DNA sequences greatly expand the available genome data from jellyfish and will be key resources for cnidarian biologists studying any of these and other phenomena.
Image: Illustration of Alatina alata, Calvadosia cruxmelitensis, and Alatina alata by: Joseph Ryan